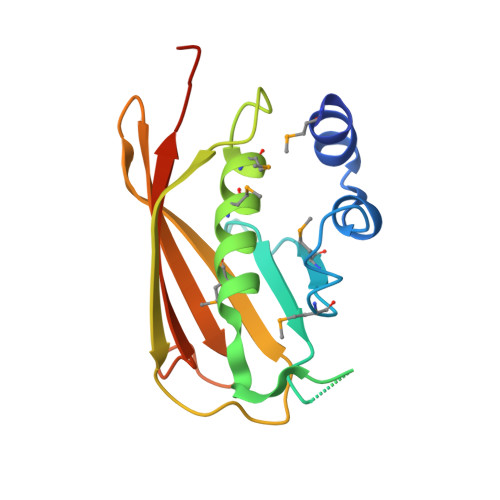
Crystal Structure of the Q7NVP2_CHRVO protein from Chromobacterium violaceum.
Vorobiev, S., Su, M., Seetharaman, J., Mao, L., Xiao, R., Ciccosanti, C., Foote, E.L., Wang, D., Everett, J.K., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.