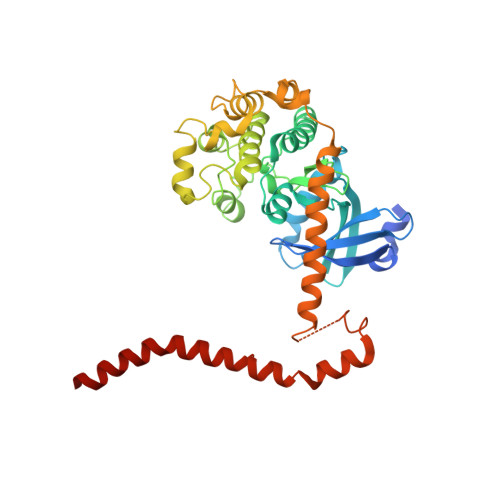
Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5.
Ni, L., Li, S., Yu, J., Min, J., Brautigam, C.A., Tomchick, D.R., Pan, D., Luo, X.(2013) Structure 21: 1757-1768
- PubMed: 23972470
- DOI: https://doi.org/10.1016/j.str.2013.07.008
- Primary Citation of Related Structures:
4LG4, 4LGD - PubMed Abstract:
The tumor-suppressive Hippo pathway controls tissue homeostasis through balancing cell proliferation and apoptosis. Activation of the kinases Mst1 and Mst2 (Mst1/2) is a key upstream event in this pathway and remains poorly understood. Mst1/2 and their critical regulators RASSFs contain Salvador/RASSF1A/Hippo (SARAH) domains that can homo- and heterodimerize. Here, we report the crystal structures of human Mst2 alone and bound to RASSF5. Mst2 undergoes activation through transautophosphorylation at its activation loop, which requires SARAH-mediated homodimerization. RASSF5 disrupts Mst2 homodimer and blocks Mst2 autoactivation. Binding of RASSF5 to already activated Mst2, however, does not inhibit its kinase activity. Thus, RASSF5 can act as an inhibitor or a potential positive regulator of Mst2, depending on whether it binds to Mst2 before or after activation-loop phosphorylation. We propose that these temporally sensitive functions of RASSFs enable the Hippo pathway to respond to and integrate diverse cellular signals.
Organizational Affiliation:
Department of Pharmacology, The University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX 75390, USA.