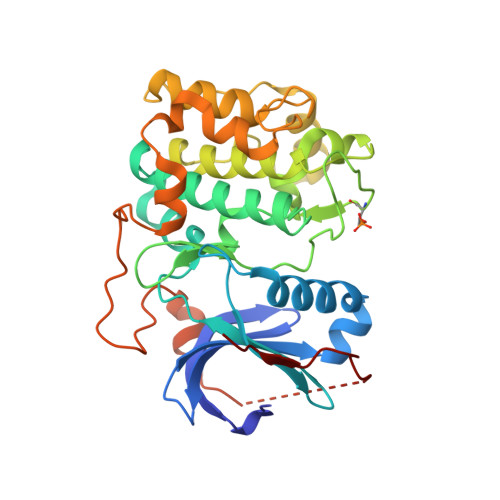
Discovery and SAR of spirochromane Akt inhibitors.
Kallan, N.C., Spencer, K.L., Blake, J.F., Xu, R., Heizer, J., Bencsik, J.R., Mitchell, I.S., Gloor, S.L., Martinson, M., Risom, T., Gross, S.D., Morales, T.H., Wu, W.I., Vigers, G.P., Brandhuber, B.J., Skelton, N.J.(2011) Bioorg Med Chem Lett 21: 2410-2414
- PubMed: 21392984
- DOI: https://doi.org/10.1016/j.bmcl.2011.02.073
- Primary Citation of Related Structures:
3QKK, 3QKL - PubMed Abstract:
A novel series of spirochromane pan-Akt inhibitors is reported. SAR optimization furnished compounds with improved enzyme potencies and excellent selectivity over the related AGC kinase PKA. Attempted replacement of the phenol hinge binder provided compounds with excellent Akt enzyme and cell activities but greatly diminished selectivity over PKA.
Organizational Affiliation:
Array BioPharma, Inc., 3200 Walnut Street, Boulder, CO 80301, USA. nkallan@arraybiopharma.com