Northeast Structural Genomics Consortium Target NmR118
Kuzin, A., Chen, Y., Seetharaman, J., Sahdev, S., Xiao, R., Ciccosanti, C., Lee, D., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein | 160 | Nitrosospira multiformis ATCC 25196 | Mutation(s): 0 Gene Names: Nmul_A1581 | 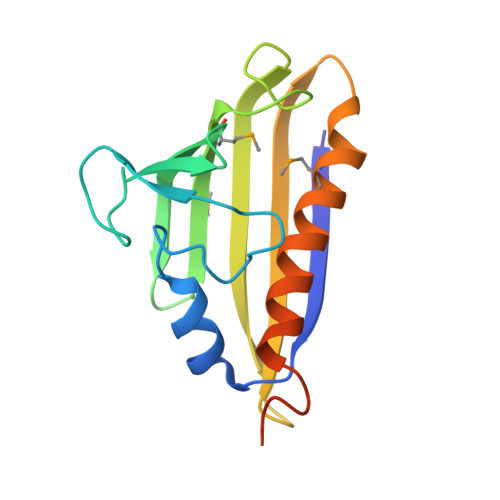 | |
UniProt | |||||
Find proteins for Q2Y8N9 (Nitrosospira multiformis (strain ATCC 25196 / NCIMB 11849 / C 71)) Explore Q2Y8N9 Go to UniProtKB: Q2Y8N9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2Y8N9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 80.409 | α = 90 |
| b = 80.409 | β = 90 |
| c = 108.256 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| AutoSol | phasing |