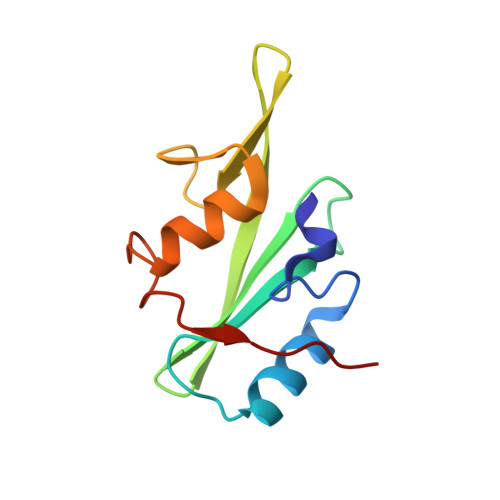
Structural Basis for the Interaction between the Growth Factor-binding Protein GRB10 and the E3 Ubiquitin Ligase NEDD4.
Huang, Q., Szebenyi, D.M.(2010) J Biol Chem 285: 42130-42139
- PubMed: 20980250
- DOI: https://doi.org/10.1074/jbc.M110.143412
- Primary Citation of Related Structures:
3M7F - PubMed Abstract:
In addition to inhibiting insulin receptor and IGF1R kinase activity by directly binding to the receptors, GRB10 can also negatively regulate insulin and IGF1 signaling by mediating insulin receptor and IGF1R degradation through ubiquitination. It has been shown that GRB10 can interact with the C2 domain of the E3 ubiquitin ligase NEDD4 through its Src homology 2 (SH2) domain. Therefore, GRB10 might act as a connector, bringing NEDD4 close to IGF1R to facilitate the ubiquitination of IGF1R by NEDD4. This is the first case in which it has been found that an SH2 domain could colocalize a ubiquitin ligase and its substrate. Here we report the crystal structure of the NEDD4 C2-GRB10 SH2 complex at 2.0 Å. The structure shows that there are three interaction interfaces between NEDD4 C2 and GRB10 SH2. The main interface centers on an antiparallel β-sheet composed of the F β-strand of GRB10 SH2 and the C β-strand of NEDD4 C2. NEDD4 C2 binds at nonclassical sites on the SH2 domain surface, far from the classical phosphotyrosine-binding pocket. Hence, this interaction is phosphotyrosine-independent, and GRB10 SH2 can bind the C2 domain of NEDD4 and the kinase domain of IGF1R simultaneously. Based on these results, a model of how NEDD4 interacts with IGF1R through GRB10 has been proposed. This report provides further evidence that SH2 domains can participate in important signaling interactions beyond the classical recognition of phosphotyrosine.
Organizational Affiliation:
MacCHESS, Cornell University, Ithaca, New York 14853, USA. qh24@cornell.edu