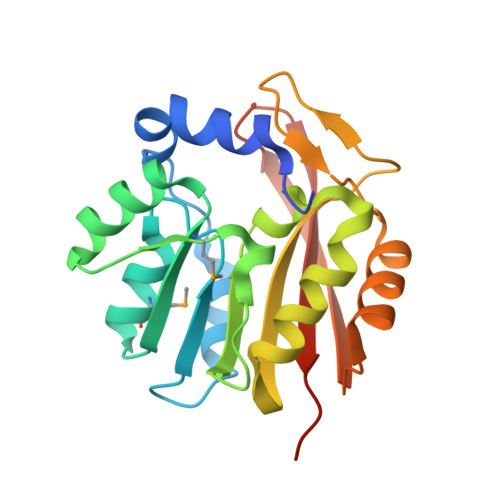
Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299
Forouhar, F., Chen, Y., Seetharaman, J., Mao, L., Xiao, R., Foote, E.L., Ciccosanti, C., Wang, H., Tong, S., K Everett, J., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.