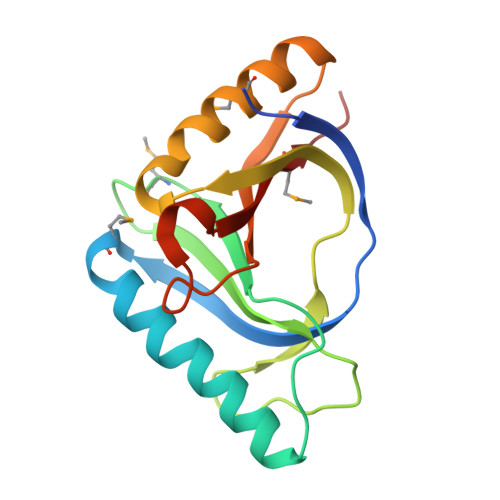
Crystal structure of an uncharacterized protein from Chlorobium tepidum. NorthEast Structural Genomics target CtR107.
Seetharaman, J., Chen, Y., Wang, H., Janjua, H., Foote, E.L., Xiao, R., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.