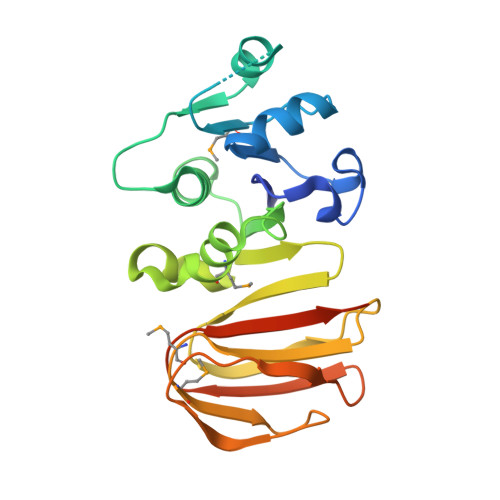
Crystal structure of the lp_1622 protein from Lactobacillus plantarum.
Vorobiev, S.M., Abashidze, M., Seetharaman, J., Zhao, L., Maglaqui, M., Foote, E.L., Ciccosanti, C., Janjua, H., Xiao, R., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.