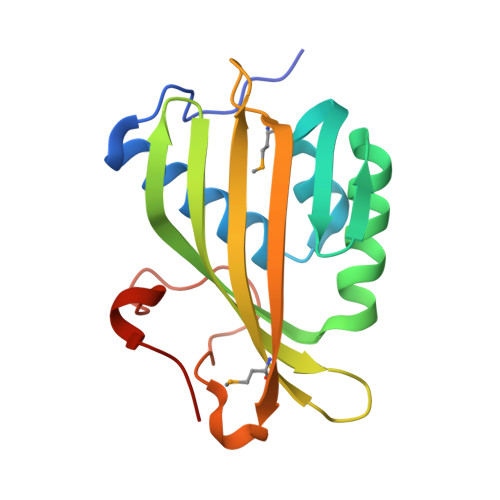
Crystal Structure of the hypothetical protein (DUF1348) from Pseudomonas fluorescens, Northeast Structural Genomics target PlR14.
Zhou, W., Forouhar, F., Seetharaman, J., Chen, C.X., Fang, Y., Cunningham, K., Ma, L.-C., Xiao, R., Liu, J., Baran, M.C., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L., Northeast Structural Genomics Consortium (NESG)To be published.