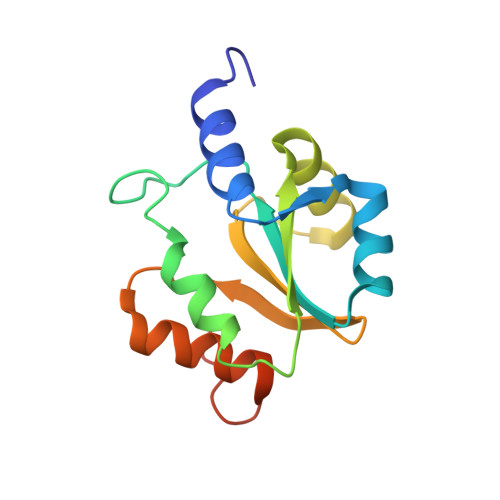
Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
Parish, D., Benach, J., Liu, G., Singarapu, K.K., Xiao, R., Acton, T., Su, M., Bansal, S., Prestegard, J.H., Hunt, J., Montelione, G.T., Szyperski, T.(2008) J Struct Funct Genomics 9: 41-49
- PubMed: 19039680
- DOI: https://doi.org/10.1007/s10969-008-9050-y
- Primary Citation of Related Structures:
2HFD, 2JZT - PubMed Abstract:
The structure of the 142-residue protein Q8ZP25_SALTY encoded in the genome of Salmonella typhimurium LT2 was determined independently by NMR and X-ray crystallography, and the structure of the 140-residue protein HYAE_ECOLI encoded in the genome of Escherichia coli was determined by NMR. The two proteins belong to Pfam (Finn et al. 34:D247-D251, 2006) PF07449, which currently comprises 50 members, and belongs itself to the 'thioredoxin-like clan'. However, protein HYAE_ECOLI and the other proteins of Pfam PF07449 do not contain the canonical Cys-X-X-Cys active site sequence motif of thioredoxin. Protein HYAE_ECOLI was previously classified as a [NiFe] hydrogenase-1 specific chaperone interacting with the twin-arginine translocation (Tat) signal peptide. The structures presented here exhibit the expected thioredoxin-like fold and support the view that members of Pfam family PF07449 specifically interact with Tat signal peptides.
Organizational Affiliation:
Department of Chemistry, Northeast Structural Genomics Consortium, The State University of New York at Buffalo, Buffalo, NY 14260, USA.