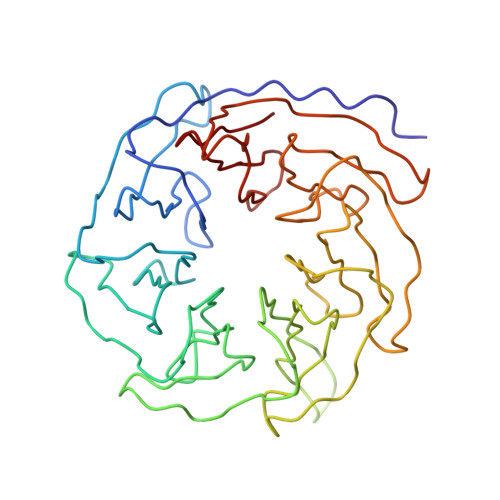
Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Sengupta, J., Nilsson, J., Gursky, R., Spahn, C.M., Nissen, P., Frank, J.(2004) Nat Struct Mol Biol 11: 957-962
- PubMed: 15334071
- DOI: https://doi.org/10.1038/nsmb822
- Primary Citation of Related Structures:
1TRJ - PubMed Abstract:
RACK1 serves as a scaffold protein for a wide range of kinases and membrane-bound receptors. It is a WD-repeat family protein and is predicted to have a beta-propeller architecture with seven blades like a Gbeta protein. Mass spectrometry studies have identified its association with the small subunit of eukaryotic ribosomes and, most recently, it has been shown to regulate initiation by recruiting protein kinase C to the 40S subunit. Here we present the results of a cryo-EM study of the 80S ribosome that positively locate RACK1 on the head region of the 40S subunit, in the immediate vicinity of the mRNA exit channel. One face of RACK1 exposes the WD-repeats as a platform for interactions with kinases and receptors. Using this platform, RACK1 can recruit other proteins to the ribosome.
Organizational Affiliation:
Health Research, Inc., Wadsworth Center, New York State Department of Health, Empire State Plaza, Albany, New York 12201-0509, USA.