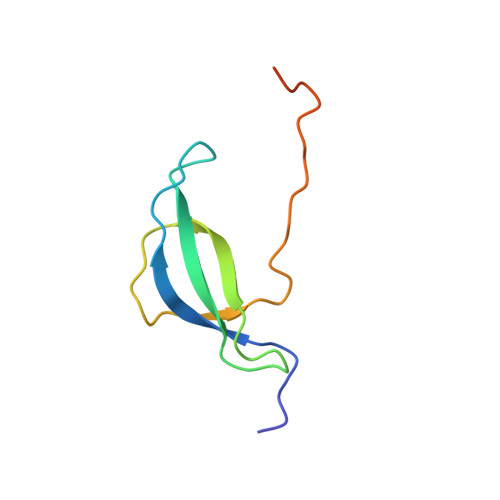
Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii.
Aramini, J.M., Huang, Y.J., Cort, J.R., Goldsmith-Fischman, S., Xiao, R., Shih, L.-Y., Ho, C.K., Liu, J., Rost, B., Honig, B., Kennedy, M.A., Acton, T.B., Montelione, G.T.(2003) Protein Sci 12: 2823-2830
- PubMed: 14627742
- DOI: https://doi.org/10.1110/ps.03359003
- Primary Citation of Related Structures:
1NY4 - PubMed Abstract:
We report NMR assignments and solution structure of the 71-residue 30S ribosomal protein S28E from the archaean Pyrococcus horikoshii, target JR19 of the Northeast Structural Genomics Consortium. The structure, determined rapidly with the aid of automated backbone resonance assignment (AutoAssign) and automated structure determination (AutoStructure) software, is characterized by a four-stranded beta-sheet with a classic Greek-key topology and an oligonucleotide/oligosaccharide beta-barrel (OB) fold. The electrostatic surface of S28E exhibits positive and negative patches on opposite sides, the former constituting a putative binding site for RNA. The 13 C-terminal residues of the protein contain a consensus sequence motif constituting the signature of the S28E protein family. Surprisingly, this C-terminal segment is unstructured in solution.
Organizational Affiliation:
Northeast Structural Genomics Consortium Center for Advanced Biotechnology and Medicine (CABM), Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, New Jersey 08854, USA.