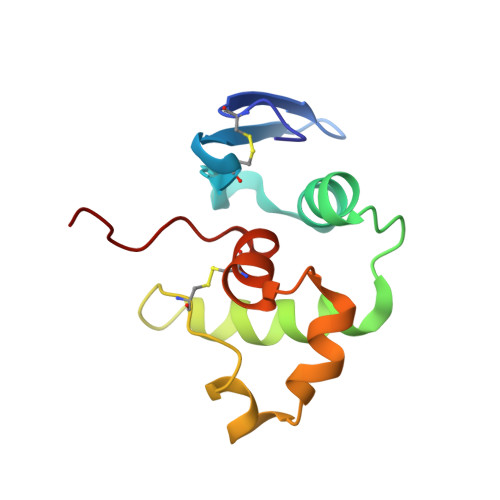
Solution structure of Pyrobaculum aerophilum DsrC, an archaeal homologue of the gamma subunit of dissimilatory sulfite reductase.
Cort, J.R., Mariappan, S.V., Kim, C.Y., Park, M.S., Peat, T.S., Waldo, G.S., Terwilliger, T.C., Kennedy, M.A.(2001) Eur J Biochem 268: 5842-5850
- PubMed: 11722571
- DOI: https://doi.org/10.1046/j.0014-2956.2001.02529.x
- Primary Citation of Related Structures:
1JI8 - PubMed Abstract:
The solution structure of DsrC, an archaeal homologue of the gamma subunit of dissimilatory sulfite reductase, has been determined by NMR spectroscopy. This 12.7-kDa protein from the hyperthermophilic archaeon Pyrobaculum aerophilum adopts a novel fold consisting of an orthogonal helical bundle with a beta hairpin along one side. A portion of the structure resembles the helix-turn-helix DNA-binding motif common in transcriptional regulator proteins. The protein contains two disulfide bonds but remains folded following reduction of the disulfides. DsrC proteins from organisms other than Pyrobaculum species do not contain these disulfide bonds. A conserved cysteine next to the C-terminus, which is not involved in the disulfide bonds, is located on a seven-residue C-terminal arm that is not part of the globular protein and is likely to dynamically sample more than one conformation.
Organizational Affiliation:
Pacific Northwest National Laboratory, Environmental Molecular Sciences Laboratory, Richland, Washington, USA.