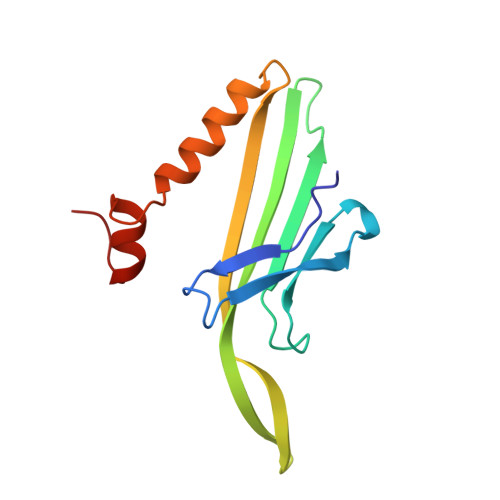
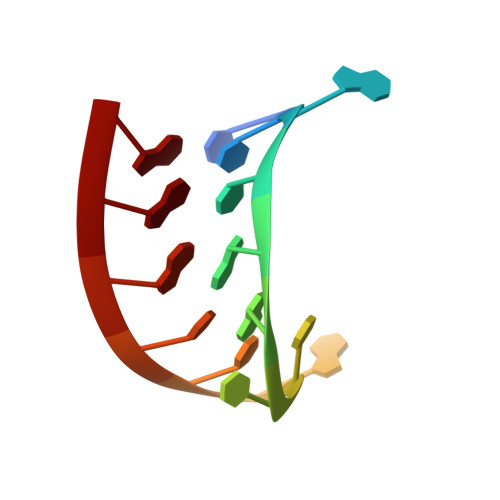
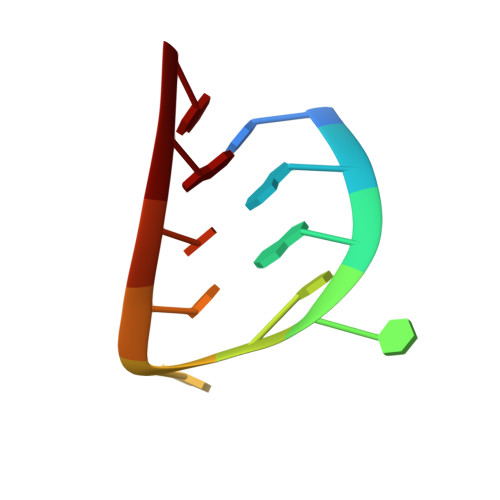
Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Convery, M.A., Rowsell, S., Stonehouse, N.J., Ellington, A.D., Hirao, I., Murray, J.B., Peabody, D.S., Phillips, S.E., Stockley, P.G.(1998) Nat Struct Biol 5: 133-139
- PubMed: 9461079
- DOI: https://doi.org/10.1038/nsb0298-133
- Primary Citation of Related Structures:
6MSF - PubMed Abstract:
The crystal structure, at 2.8 A resolution, of an RNA aptamer bound to bacteriophage MS2 coat protein has been determined. It provides an opportunity to compare the interactions of MS2 coat protein and wild type operator with those of an aptamer, whose secondary structure differs from the wild type RNA in having a three-base loop (compared to a tetraloop) and an additional base pair between this loop and the sequence-specific recognition element in the stem. The RNA binds in the same location on the coat protein as the wild type operator and maintains many of the same RNA-protein interactions. In order to achieve this, the RNA stem loop undergoes a concerted rearrangement of the 3' side while leaving the 5' side and the loop interactions largely unchanged, illustrating the ability of RNA to present similar molecular recognition surfaces from distinct primary and secondary structures.
Organizational Affiliation:
School of Biochemistry and Molecular Biology, North of England Structural Biology Centre, University of Leeds, UK.