A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
Liberman, J.A., Guo, M., Jenkins, J.L., Krucinska, J., Chen, Y., Carey, P.R., Wedekind, J.E.(2012) J Am Chem Soc 134: 16933-16936
- PubMed: 22989273
- DOI: https://doi.org/10.1021/ja3070528
- Primary Citation of Related Structures:
4G6P, 4G6R, 4G6S - PubMed Abstract:
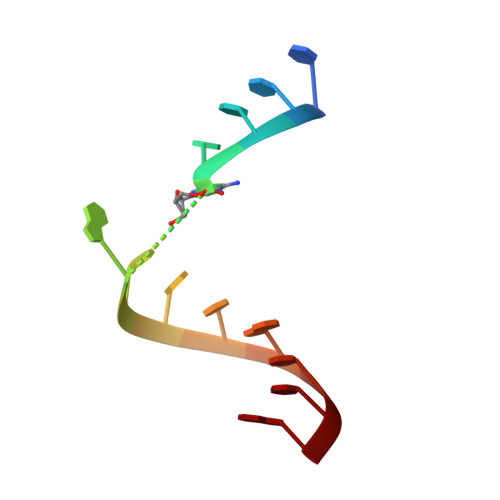
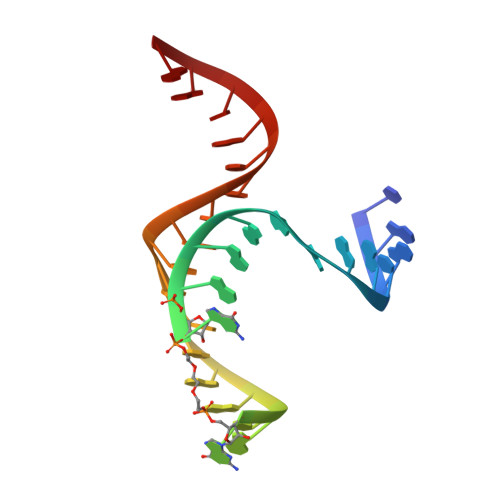
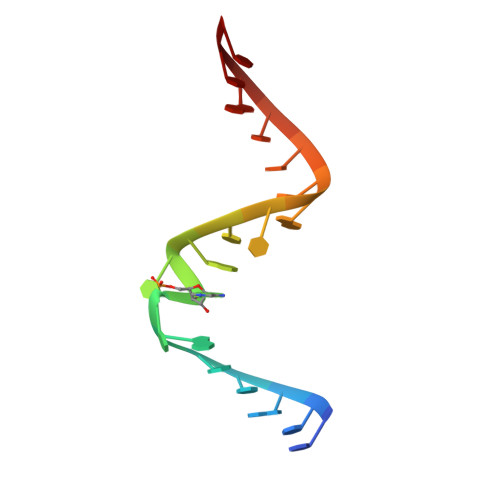
One mechanism by which ribozymes can accelerate biological reactions is by adopting folds that favorably perturb nucleobase ionization. Herein we used Raman crystallography to directly measure pK(a) values for the Ade38 N1 imino group of a hairpin ribozyme in distinct conformational states. A transition-state analogue gave a pK(a) value of 6.27 ± 0.05, which agrees strikingly well with values measured by pH-rate analyses. To identify the chemical attributes that contribute to the shifted pK(a), we determined crystal structures of hairpin ribozyme variants containing single-atom substitutions at the active site and measured their respective Ade38 N1 pK(a) values. This approach led to the identification of a single interaction in the transition-state conformation that elevates the base pK(a) > 0.8 log unit relative to the precatalytic state. The agreement of the microscopic and macroscopic pK(a) values and the accompanying structural analysis supports a mechanism in which Ade38 N1(H)+ functions as a general acid in phosphodiester bond cleavage. Overall the results quantify the contribution of a single electrostatic interaction to base ionization, which has broad relevance for understanding how RNA structure can control chemical reactivity.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Center for RNA Biology, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Box 712, Rochester, New York 14642, USA.