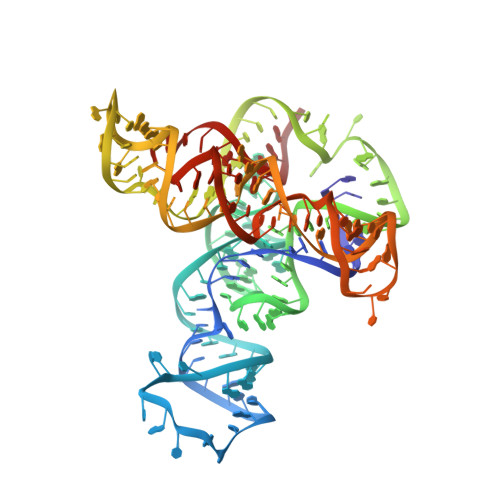
Structural basis of cooperative ligand binding by the glycine riboswitch.
Butler, E.B., Xiong, Y., Wang, J., Strobel, S.A.(2011) Chem Biol 18: 293-298
- PubMed: 21439473
- DOI: https://doi.org/10.1016/j.chembiol.2011.01.013
- Primary Citation of Related Structures:
3P49 - PubMed Abstract:
The glycine riboswitch regulates gene expression through the cooperative recognition of its amino acid ligand by a tandem pair of aptamers. A 3.6 Å crystal structure of the tandem riboswitch from the glycine permease operon of Fusobacterium nucleatum reveals the glycine binding sites and an extensive network of interactions, largely mediated by asymmetric A-minor contacts, that serve to communicate ligand binding status between the aptamers. These interactions provide a structural basis for how the glycine riboswitch cooperatively regulates gene expression.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA.