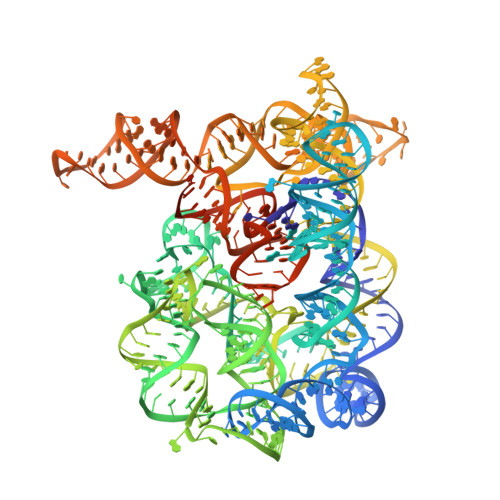
Tertiary architecture of the Oceanobacillus iheyensis group II intron.
Toor, N., Keating, K.S., Fedorova, O., Rajashankar, K., Wang, J., Pyle, A.M.(2010) RNA 16: 57-69
- PubMed: 19952115
- DOI: https://doi.org/10.1261/rna.1844010
- Primary Citation of Related Structures:
3IGI - PubMed Abstract:
Group II introns are large ribozymes that act as self-splicing and retrotransposable RNA molecules. They are of great interest because of their potential evolutionary relationship to the eukaryotic spliceosome, their continued influence on the organization of many genomes in bacteria and eukaryotes, and their potential utility as tools for gene therapy and biotechnology. One of the most interesting features of group II introns is their relative lack of nucleobase conservation and covariation, which has long suggested that group II intron structures are stabilized by numerous unusual tertiary interactions and backbone-mediated contacts. Here, we provide a detailed description of the tertiary interaction networks within the Oceanobacillus iheyensis group IIC intron, for which a crystal structure was recently solved to 3.1 A resolution. The structure can be described as a set of several intricately constructed tertiary interaction nodes, each of which contains a core of extended stacking networks and elaborate motifs. Many of these nodes are surrounded by a web of ribose zippers, which appear to further stabilize local structure. As predicted from biochemical and genetic studies, the group II intron provides a wealth of new information on strategies for RNA folding and tertiary structural organization.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06511, USA.