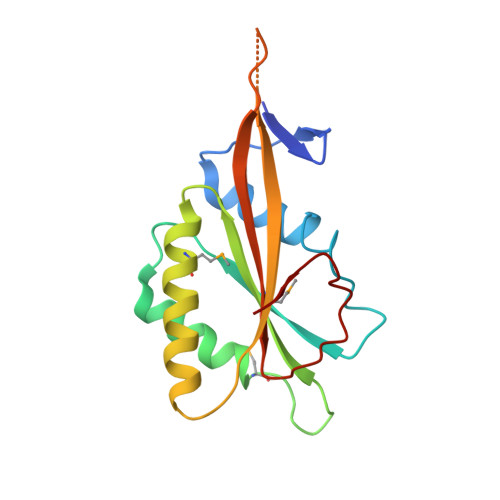
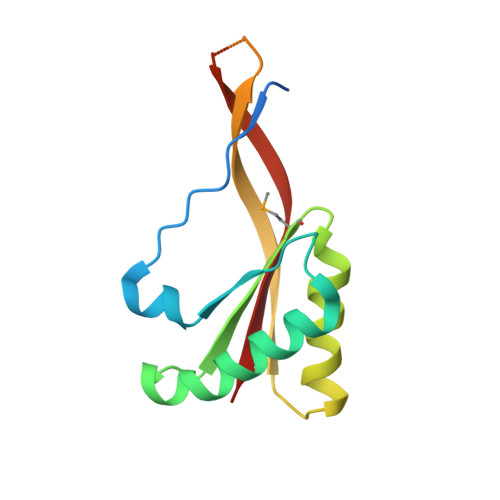
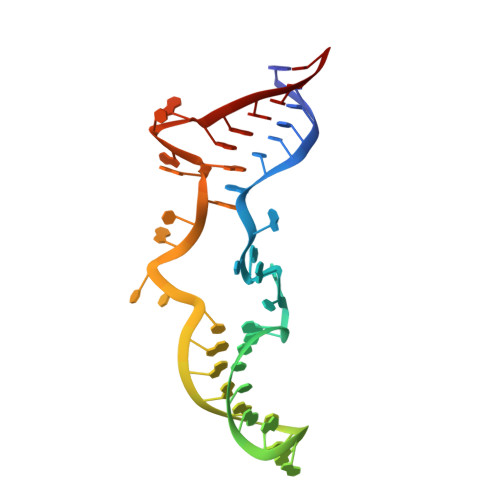
Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain
Perederina, A., Esakova, O., Quan, C., Khanova, E., Krasilnikov, A.S.(2010) EMBO J 29: 761-769
- PubMed: 20075859
- DOI: https://doi.org/10.1038/emboj.2009.396
- Primary Citation of Related Structures:
3IAB - PubMed Abstract:
Ribonuclease (RNase) P is a site-specific endoribonuclease found in all kingdoms of life. Typical RNase P consists of a catalytic RNA component and a protein moiety. In the eukaryotes, the RNase P lineage has split into two, giving rise to a closely related enzyme, RNase MRP, which has similar components but has evolved to have different specificities. The eukaryotic RNases P/MRP have acquired an essential helix-loop-helix protein-binding RNA domain P3 that has an important function in eukaryotic enzymes and distinguishes them from bacterial and archaeal RNases P. Here, we present a crystal structure of the P3 RNA domain from Saccharomyces cerevisiae RNase MRP in a complex with RNase P/MRP proteins Pop6 and Pop7 solved to 2.7 A. The structure suggests similar structural organization of the P3 RNA domains in RNases P/MRP and possible functions of the P3 domains and proteins bound to them in the stabilization of the holoenzymes' structures as well as in interactions with substrates. It provides the first insight into the structural organization of the eukaryotic enzymes of the RNase P/MRP family.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, PA 16802, USA.