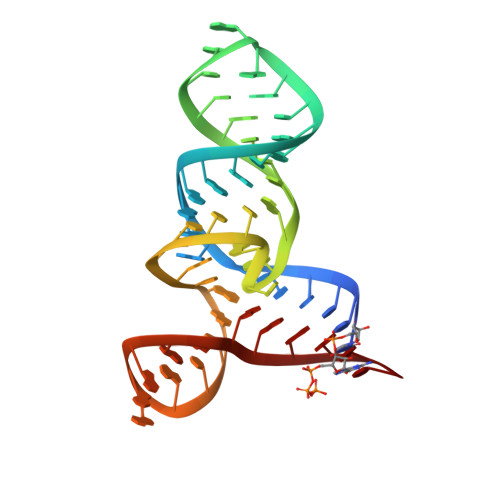
Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Lu, C., Smith, A.M., Fuchs, R.T., Ding, F., Rajashankar, K., Henkin, T.M., Ke, A.(2008) Nat Struct Mol Biol 15: 1076-1083
- PubMed: 18806797
- DOI: https://doi.org/10.1038/nsmb.1494
- Primary Citation of Related Structures:
3E5C, 3E5E, 3E5F - PubMed Abstract:
Three distinct classes of S-adenosyl-L-methionine (SAM)-responsive riboswitches have been identified that regulate bacterial gene expression at the levels of transcription attenuation or translation inhibition. The S(MK) box (SAM-III) translational riboswitch has been identified in the SAM synthetase gene in members of the Lactobacillales. Here we report the 2.2-A crystal structure of the Enterococcus faecalis S(MK) box riboswitch. The Y-shaped riboswitch organizes its conserved nucleotides around a three-way junction for SAM recognition. The Shine-Dalgarno sequence, which is sequestered by base-pairing with the anti-Shine-Dalgarno sequence in response to SAM binding, also directly participates in SAM recognition. The riboswitch makes extensive interactions with the adenosine and sulfonium moieties of SAM but does not appear to recognize the tail of the methionine moiety. We captured a structural snapshot of the S(MK) box riboswitch sampling the near-cognate ligand S-adenosyl-L-homocysteine (SAH) in which SAH was found to adopt an alternative conformation and fails to make several key interactions.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Cornell University, 251 Biotechnology Building, Ithaca, New York 14853, USA.