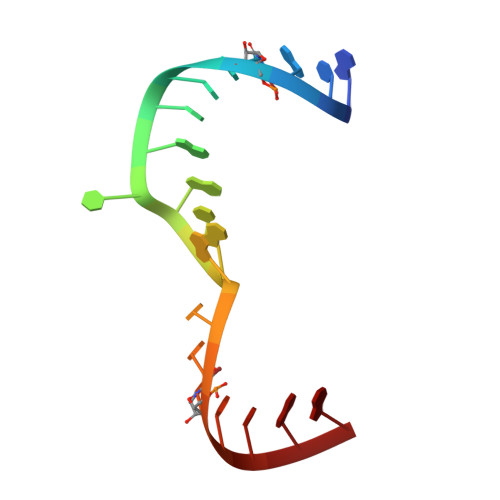
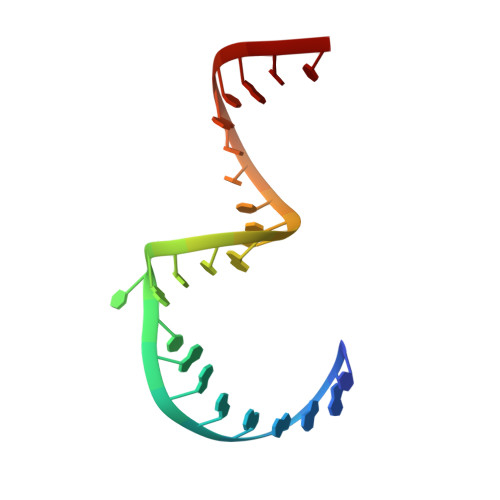Structure of hepatitis C virus IRES subdomain IIa.
Zhao, Q., Han, Q., Kissinger, C.R., Hermann, T., Thompson, P.A.(2008) Acta Crystallogr D Biol Crystallogr 64: 436-443
- PubMed: 18391410
- DOI: https://doi.org/10.1107/S0907444908002011
- Primary Citation of Related Structures:
2PN3, 2PN4 - PubMed Abstract:
The hepatitis C (HCV) internal ribosome entry site (IRES) element plays a central role in cap-independent translation of the viral genomic RNA. The unique conformation of IRES domain II is critical for 80S ribosomal assembly and initiation of viral translation. Here, the crystal structure of subdomain IIa of the HCV IRES has been determined at 2.3 A resolution, revealing the positions of divalent metal ions and complex inter-strand interactions that stabilize the L-shaped conformation of the RNA. The presence of divalent metal ions was necessary for crystal formation. Magnesium ions occupy specific sites that appear to be critical for the formation of the folded conformation. Subdomain IIa also was crystallized in the presence of strontium, which improved the diffraction quality of the crystals and the ability to identify interactions of the RNA with metal ions and tightly bound water molecules. The hinge region and noncanonical G-U base-pair motifs are stabilized by divalent metal ions and provide unique structural features that are potential interaction sites for small-molecule ligands. The information obtained from the crystal structure provides a basis for structure-guided design of HCV translation inhibitors targeting disruption of ribosomal assembly.
Organizational Affiliation:
Department of Structural Chemistry, Anadys Pharmaceuticals Inc., 3115 Merryfield Row, San Diego, CA 92121, USA. qiangzhao@hotmail.com