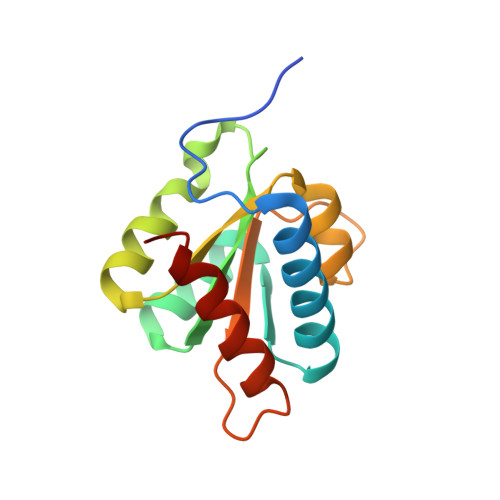
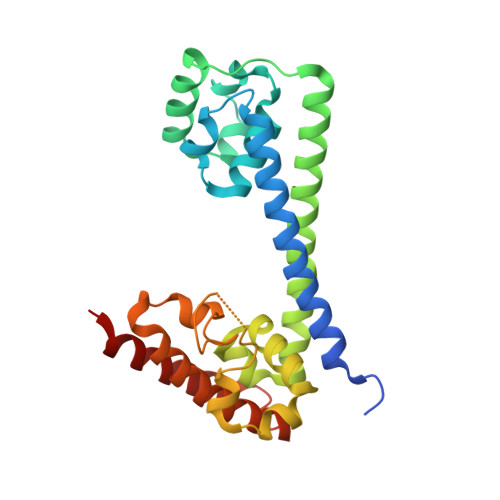
Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP.
Liu, S., Li, P., Dybkov, O., Nottrott, S., Hartmuth, K., Luhrmann, R., Carlomagno, T., Wahl, M.C.(2007) Science 316: 115-120
- PubMed: 17412961
- DOI: https://doi.org/10.1126/science.1137924
- Primary Citation of Related Structures:
2OZB - PubMed Abstract:
Although highly homologous, the spliceosomal hPrp31 and the nucleolar Nop56 and Nop58 (Nop56/58) proteins recognize different ribonucleoprotein (RNP) particles. hPrp31 interacts with complexes containing the 15.5K protein and U4 or U4atac small nuclear RNA (snRNA), whereas Nop56/58 associate with 15.5K-box C/D small nucleolar RNA complexes. We present structural and biochemical analyses of hPrp31-15.5K-U4 snRNA complexes that show how the conserved Nop domain in hPrp31 maintains high RNP binding selectivity despite relaxed RNA sequence requirements. The Nop domain is a genuine RNP binding module, exhibiting RNA and protein binding surfaces. Yeast two-hybrid analyses suggest a link between retinitis pigmentosa and an aberrant hPrp31-hPrp6 interaction that blocks U4/U6-U5 tri-snRNP formation.
Organizational Affiliation:
Abteilung Zelluläre Biochemie, Max-Planck-Institut für Biophysikalische Chemie, Am Fassberg 11, D-37077 Göttingen, Germany.