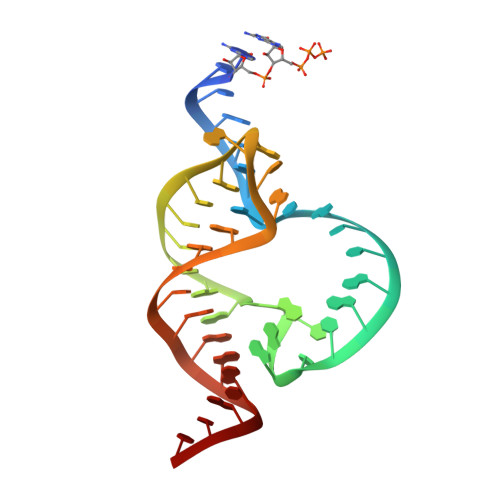
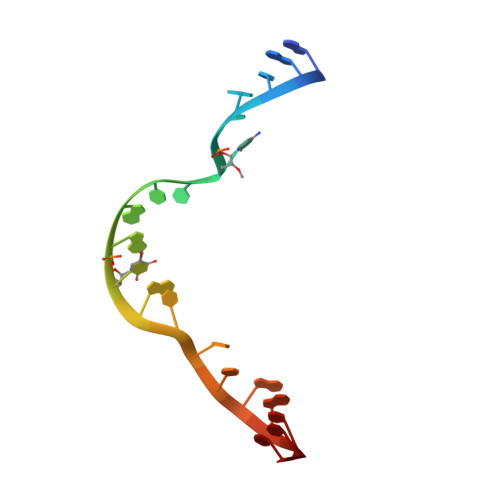
Solvent structure and hammerhead ribozyme catalysis.
Martick, M., Lee, T.S., York, D.M., Scott, W.G.(2008) Chem Biol 15: 332-342
- PubMed: 18420140
- DOI: https://doi.org/10.1016/j.chembiol.2008.03.010
- Primary Citation of Related Structures:
2OEU - PubMed Abstract:
Although the hammerhead ribozyme is regarded as a prototype for understanding RNA catalysis, the mechanistic roles of associated metal ions and water molecules in the cleavage reaction remain controversial. We have investigated the catalytic potential of observed divalent metal ions and water molecules bound to a 2 A structure of the full-length hammerhead ribozyme by using X-ray crystallography in combination with molecular dynamics simulations. A single Mn(2+) is observed to bind directly to the A9 phosphate in the active site, accompanying a hydrogen-bond network involving a well-ordered water molecule spanning N1 of G12 (the general base) and 2'-O of G8 (previously implicated in general acid catalysis) that we propose, based on molecular dynamics calculations, facilitates proton transfer in the cleavage reaction. Phosphate-bridging metal interactions and other mechanistic hypotheses are also tested with this approach.
Organizational Affiliation:
Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.