Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Zuo, X., Wang, J., Yu, P., Eyler, D., Xu, H., Starich, M.R., Tiede, D.M., Simon, A.E., Kasprzak, W., Schwieters, C.D., Shapiro, B.A., Wang, Y.X.(2010) Proc Natl Acad Sci U S A 107: 1385-1390
- PubMed: 20080629
- DOI: https://doi.org/10.1073/pnas.0908140107
- Primary Citation of Related Structures:
2KRL - PubMed Abstract:
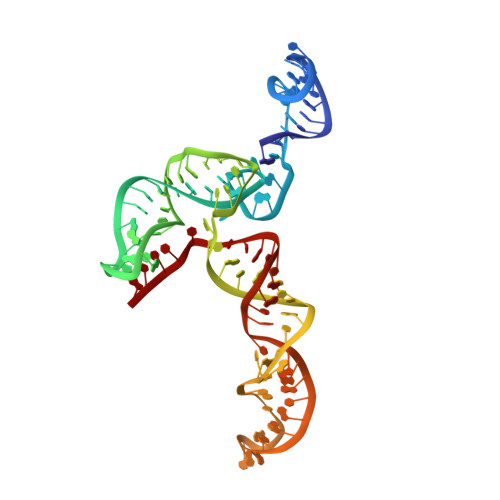
The 3(') untranslated region (3(') UTR) of turnip crinkle virus (TCV) genomic RNA contains a cap-independent translation element (CITE), which includes a ribosome-binding structural element (RBSE) that participates in recruitment of the large ribosomal subunit. In addition, a large symmetric loop in the RBSE plays a key role in coordinating the incompatible processes of viral translation and replication, which require enzyme progression in opposite directions on the viral template. To understand the structural basis for the large ribosomal subunit recruitment and the intricate interplay among different parts of the molecule, we determined the global structure of the 102-nt RBSE RNA using solution NMR and small-angle x-ray scattering. This RNA has many structural features that resemble those of a tRNA in solution. The hairpins H1 and H2, linked by a 7-nucleotide linker, form the upper part of RBSE and hairpin H3 is relatively independent from the rest of the structure and is accessible to interactions. This global structure provides insights into the three-dimensional layout for ribosome binding, which may serve as a structural basis for its involvement in recruitment of the large ribosomal subunit and the switch between viral translation and replication. The experimentally determined three-dimensional structure of a functional element in the 3(') UTR of an RNA from any organism has not been previously reported. The RBSE structure represents a prototype structure of a new class of RNA structural elements involved in viral translation/replication processes.
Organizational Affiliation:
Protein Nucleic Acid Interaction Section, Structural Biophysics Laboratory, National Cancer Institute at Frederick, National Institutes of Health, Frederick, MD 21702, USA.