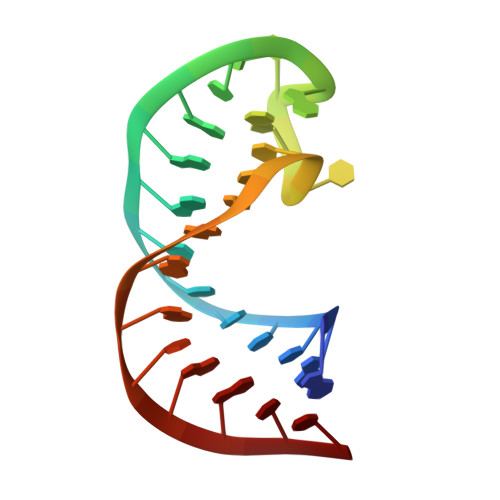
Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Flodell, S., Petersen, M., Girard, F., Zdunek, J., Kidd-Ljunggren, K., Schleucher, J., Wijmenga, S.(2006) Nucleic Acids Res 34: 4449-4457
- PubMed: 16945960
- DOI: https://doi.org/10.1093/nar/gkl582
- Primary Citation of Related Structures:
2IXY, 2IXZ - PubMed Abstract:
Hepatitis B virus (HBV) replication is initiated by HBV RT binding to the highly conserved encapsidation signal, epsilon, at the 5' end of the RNA pregenome. Epsilon contains an apical stem-loop, whose residues are either totally conserved or show rare non-disruptive mutations. Here we present the structure of the apical stem-loop based on NOE, RDC and (1)H chemical shift NMR data. The (1)H chemical shifts proved to be crucial to define the loop conformation. The loop sequence 5'-CUGUGC-3' folds into a UGU triloop with a CG closing base pair and a bulged out C and hence forms a pseudo-triloop, a proposed protein recognition motif. In the UGU loop conformations most consistent with experimental data, the guanine nucleobase is located on the minor groove face and the two uracil bases on the major groove face. The underlying helix is disrupted by a conserved non-paired U bulge. This U bulge adopts multiple conformations, with the nucleobase being located either in the major groove or partially intercalated in the helix from the minor groove side, and bends the helical stem. The pseudo-triloop motif, together with the U bulge, may represent important anchor points for the initial recognition of epsilon by the viral RT.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Umeå University, 901 87 Umeå, Sweden.