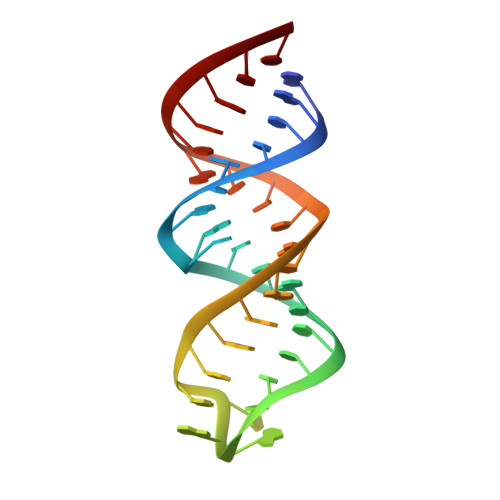
Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2
Nomura, Y., Kajikawa, M., Baba, S., Nakazato, S., Imai, T., Sakamoto, T., Okada, N., Kawai, G.(2006) Nucleic Acids Res 34: 5184-5193
- PubMed: 17000640
- DOI: https://doi.org/10.1093/nar/gkl664
- Primary Citation of Related Structures:
2FDT - PubMed Abstract:
The eel long interspersed element (LINE) UnaL2 and its partner short interspersed element (SINE) share a conserved 3' tail that is critical for their retrotransposition. The predicted secondary structure of the conserved 3' tail of UnaL2 RNA contains a stem region with a putative internal loop. Deletion of the putative internal loop region abolishes UnaL2 mobilization, indicating that this putative internal loop is required for UnaL2 retrotransposition; the exact role of the putative internal loop in retrotransposition, however, has not been elucidated. To establish a structure-based foundation on which to address the issue of the putative internal loop function in retrotransposition, we used NMR to determine the solution structure of a 36 nt RNA derived from the 3' conserved tail of UnaL2. The region forms a compact structure containing a single bulged cytidine and a U-U mismatch. The bulge and mismatch region have conformational flexibility and molecular dynamics simulation indicate that the entire stem of the 3' conserved tail RNA can anisotropically fluctuate at the bulge and mismatch region. Our structural and mutational analyses suggest that stem flexibility contributes to UnaL2 function and that the bulged cytidine and the U-U mismatch are required for efficient retrotransposition.
Organizational Affiliation:
Department of Life and Environmental Sciences, Faculty of Engineering, Chiba Institute of Technology, 2-17-1 Tsudanuma, Narashino, Chiba 275-0016, Japan.