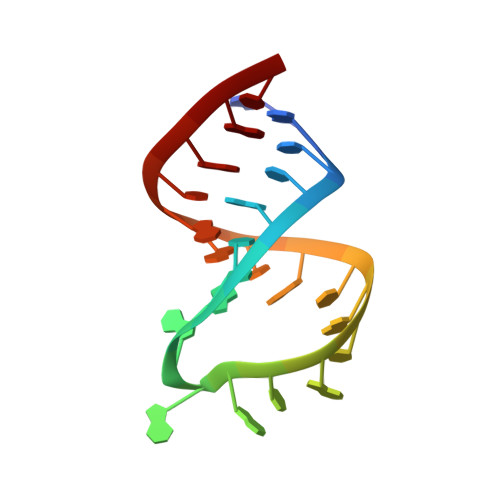
A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer
Kieken, F., Paquet, F., Brule, F., Paoletti, J., Lancelot, G.(2006) Nucleic Acids Res 34: 343-352
- PubMed: 16410614
- DOI: https://doi.org/10.1093/nar/gkj427
- Primary Citation of Related Structures:
2F4X - PubMed Abstract:
Dimerization of genomic RNA is directly related with the event of encapsidation and maturation of the virion. The initiating sequence of the dimerization is a short autocomplementary region in the hairpin loop SL1. We describe here a new solution structure of the RNA dimerization initiation site (DIS) of HIV-1(Lai). NMR pulsed field-gradient spin-echo techniques and multidimensional heteronuclear NMR spectroscopy indicate that this structure is formed by two hairpins linked by six Watson-Crick GC base pairs. Hinges between the stems and the loops are stabilized by intra and intermolecular interactions involving the A8, A9 and A16 adenines. The coaxial alignment of the three A-type helices present in the structure is supported by previous crystallography analysis but the A8 and A9 adenines are found in a bulged in position. These data suggest the existence of an equilibrium between bulged in and bulged out conformations in solution.
Organizational Affiliation:
Centre de Biophysique Moléculaire, Orléans University, Rue Charles Sadron, 45071 Orléans Cedex2, France.