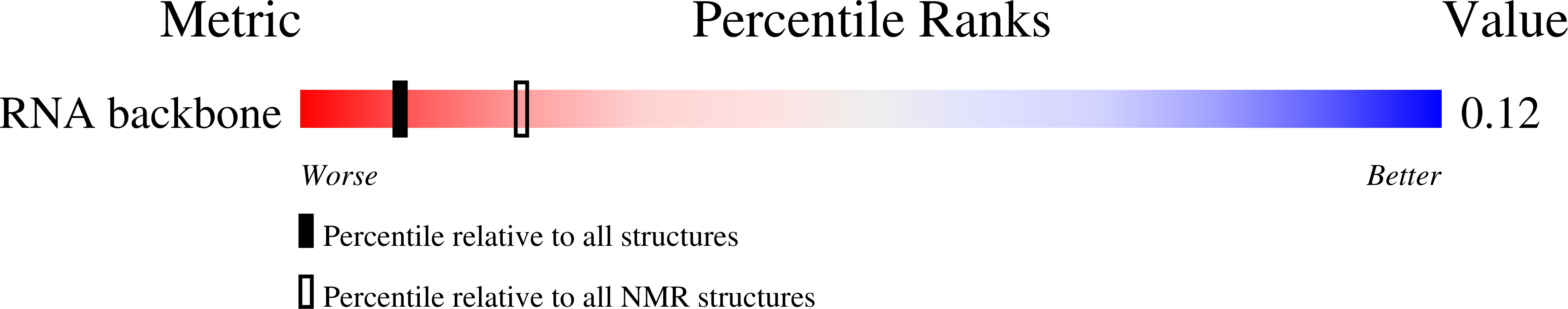The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical
Cornish, P.V., Stammler, S.N., Giedroc, D.P.(2006) RNA 12: 1959-1969
- PubMed: 17000902
- DOI: https://doi.org/10.1261/rna.199006
- Primary Citation of Related Structures:
2AP0, 2AP5 - PubMed Abstract:
The helical junction region of a -1 frameshift stimulating hairpin-type mRNA pseudoknot from sugarcane yellow leaf virus (ScYLV) is characterized by a novel C27.(G7-C14) loop 2-stem 1 minor groove base triple, which is stacked on a C8+.(G12-C28) loop 1-stem 2 major groove base triple. Substitution of C27 with adenosine reduces frameshifting efficiency to a level just twofold above the slip-site alone. Here, we show that the global structure of the C27A ScYLV RNA is nearly indistinguishable from the wild-type counterpart, despite the fact that the helical junction region is altered and incorporates the anticipated isostructural A27.(G7-C14) minor groove base triple. This interaction mediates a 2.3-A displacement of C8+ driven by an A27 N6-C8+ O2 hydrogen bond as part of an A(n-1).C+.G-Cn base quadruple. The helical junction regions of the C27A ScYLV and the beet western yellows virus (BWYV) pseudoknots are essentially superimposable, the latter of which contains an analogous A25.(G7-C14) minor groove base triple. These results reveal that the global ground-state structure is not strongly correlated with frameshift stimulation and point to a reduced thermodynamic stability and/or enhanced kinetic lability that derives from an altered helical junction architecture in the C27A ScYLV RNA as a significant determinant for setting frameshifting efficiencies in plant luteoviral mRNA pseudoknots.
Organizational Affiliation:
Department of Biochemistry and Biophysics, 2128 TAMU, Texas A&M University, College Station, Texas 77843-2128, USA.