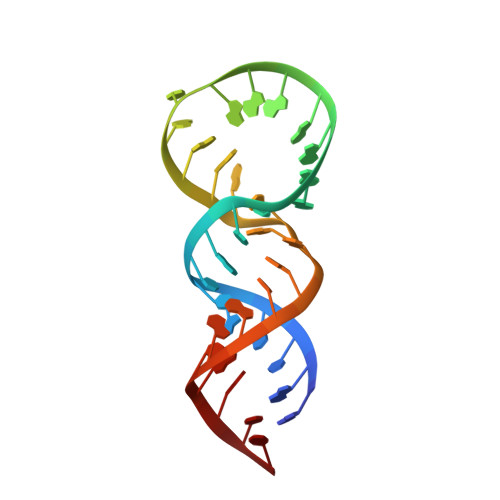
High resolution structure of a picornaviral internal cis-acting replication element(cre).
Thiviyanathan, V., Yang, Y., Kaluarachchi, K., Reynbrand, R., Gorenstein, D.G., Lemon, S.M.(2004) Proc Natl Acad Sci U S A 101: 12688-12693
- PubMed: 15314212
- DOI: https://doi.org/10.1073/pnas.0403079101
- Primary Citation of Related Structures:
1T28 - PubMed Abstract:
Picornaviruses constitute a medically important family of RNA viruses in which genome replication critically depends on a small RNA element, the cis-acting replication element (cre), that templates 3D(pol) polymerase-catalyzed uridylylation of the protein primer for RNA synthesis, VPg. We report the solution structure of the 33-nt cre of human rhinovirus 14 under solution conditions optimal for uridylylation in vitro. The cre adopts a stem-loop conformation with an extended duplex stem supporting a novel 14-nt loop that derives stability from base-stacking interactions. Base-pair interactions are absent within the loop, and base substitutions within the loop that favor such interactions are detrimental to viral RNA replication. Conserved adenosines in the 5' loop sequence that participate in a slide-back mechanism of VPg-pUpU synthesis are oriented to the inside of the loop but are available for base templating during uridylation. The structure explains why substitutions of the 3' loop nucleotides have little impact on conformation of the critical 5' loop bases and accounts for wide variation in the sequences of cres from different enteroviruses and rhinoviruses.
Organizational Affiliation:
Sealy Center for Structural Biology, University of Texas Medical Branch, Galveston, TX 77555, USA.