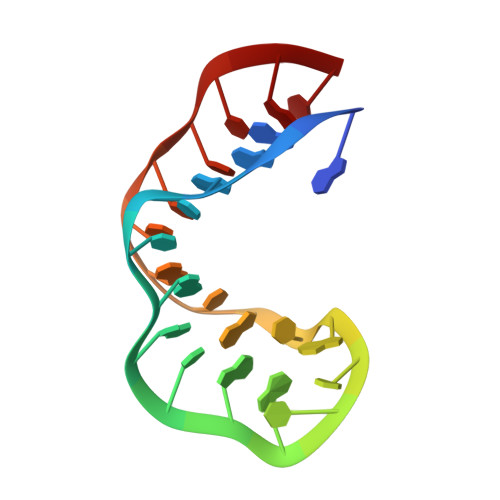
Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Borer, P.N., Lin, Y., Wang, S., Roggenbuck, M.W., Gott, J.M., Uhlenbeck, O.C., Pelczer, I.(1995) Biochemistry 34: 6488-6503
- PubMed: 7756280
- DOI: https://doi.org/10.1021/bi00019a030
- Primary Citation of Related Structures:
1RHT - PubMed Abstract:
The three-dimensional conformation of a 24-nucleotide variant of the RNA binding sequence for the coat protein of bacteriophage R17 has been analyzed using NMR, molecular dynamics, and energy minimization. The imino proton spectrum is consistent with base pairing requirements for coat protein binding known from biochemical studies. All 185 of the nonexchangeable protons were assigned using a variety of homonuclear 2D and 3D NMR methods. Measurements of nuclear Overhauser enhancements and two-quantum correlations were made at 500 MHz. New procedures were developed to characterize as many resonances as possible, including deconvolution and path analysis methods. An average of 21 distance constraints per residue were used in molecular dynamics calculations to obtain preliminary folded structures for residues 3-21. The unpaired A8 residue is stacked in the stem, and the entire region from G7 to C15 in the upper stem and loop appears to be flexible. Several of these residues have a large fraction of S-puckered ribose rings, rather than the N-forms characteristic of RNA duplexes. There is considerable variation in the low-energy loop conformations that satisfy the distance constraints at this preliminary level of refinement. The Shine-Dalgarno ribosome binding site is exposed, and only two apparently weak base pairs would have to break for the 16S ribosomal RNA to bind and the ribosome to initiate translation of the replicase gene. Although the loop form must be regarded as tentative, the known interaction sites with the coat protein are easily accessible from the major groove side of the loop.
Organizational Affiliation:
Chemistry Department, Syracuse University, New York 13244-4100, USA.