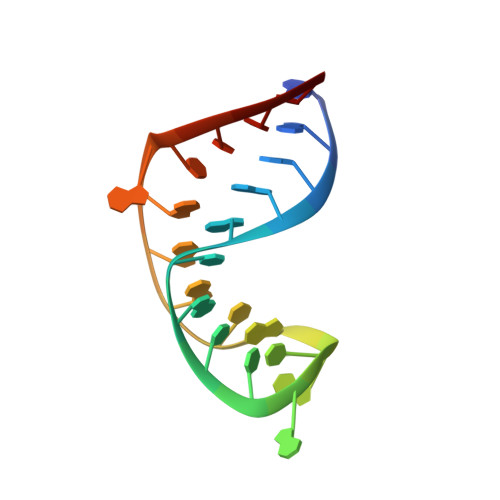
Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Yuan, Y., Kerwood, D.J., Paoletti, A.C., Shubsda, M.F., Borer, P.N.(2003) Biochemistry 42: 5259-5269
- PubMed: 12731867
- DOI: https://doi.org/10.1021/bi034084a
- Primary Citation of Related Structures:
1OSW - PubMed Abstract:
The 5'-leader of HIV-1 RNA controls many viral functions. Nucleocapsid (NC) domains of gag-precursor proteins select genomic RNA for packaging by binding several sites in the leader. One is likely to be a stem defect in SL1 that can adopt either a 1 x 3 internal loop, SL1i (including G247, A271, G272, G273) or a 1 x 1 internal loop (G247 x G273) near a two-base bulge (A269-G270). It is likely that these two conformations are both present and exchange readily. A 23mer RNA construct described here models SL1i and cannot slip into the alternate form. It forms a 1:1 complex with NCp7, which interacts most strongly at G247 and G272 (K(d) = 140 nM). This demonstrates that a linear G-X-G sequence is unnecessary for high-affinity binding. The NMR-based structure shows an easily broken G247:A271 base pair. G247 stacks on both of its immediate neighbors and A271 on its 5'-neighbor; G272 and G273 are partially ordered. A bend in the helix axis between the SL1 stems on either side of the internal loop is probable. An important step in maturation of the virus is the transition from an apical loop-loop interaction to a dimer involving intermolecular interactions along the full length of SL1. A bend in the stem may be important in relieving strain and ensuring that the strands do not become entangled during the transition. A stem defect with special affinity for NCp7 may accelerate the rate of the dimer transformation. This complex could become an important target for anti-HIV drug development, where a drug could exert its action near a high-energy intermediate on the pathway for maturation of the dimer.
Organizational Affiliation:
Department of Chemistry, Graduate Program in Structural Biology, Biochemistry, and Biophysics, Syracuse University, Syracuse, New York 13244-4100, USA.