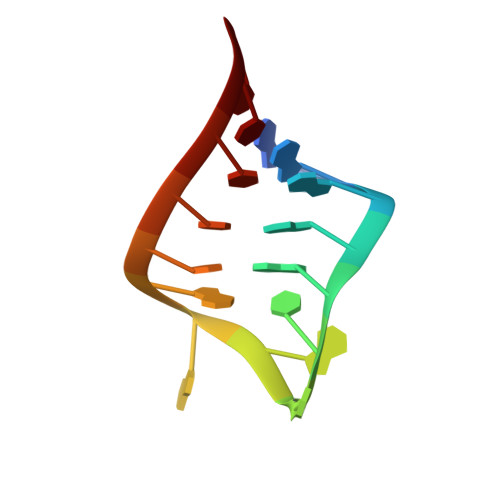
The solution structure of an essential stem-loop of human telomerase RNA
Leeper, T., Leulliot, N., Varani, G.(2003) Nucleic Acids Res 31: 2614-2621
- PubMed: 12736311
- DOI: https://doi.org/10.1093/nar/gkg351
- Primary Citation of Related Structures:
1OQ0 - PubMed Abstract:
The ribonucleoprotein enzyme telomerase maintains chromosome ends in most eukaryotes and is critical for a cell's genetic stability and its proliferative viability. All telomerases contain a catalytic protein component homologous to viral reverse transcriptases (TERT) and an RNA (TR) that provides the template sequence as well as a scaffold for ribonucleoprotein assembly. Vertebrate telomerase RNAs have three essential domains: the template, activation and stability domains. Here we report the NMR structure of an essential RNA element derived from the human telomerase RNA activation domain. The sequence forms a stem-loop structure stabilized by a GU wobble pair formed by two of the five unpaired residues capping a short double helical region. The remaining three loop residues are in a well-defined conformation and form phosphate-base stacking interactions reminiscent of other RNA loop structures. Mutations of these unpaired nucleotides abolish enzymatic activity. The structure rationalizes a number of biochemical observations, and allows us to propose how the loop may function in the telomerase catalytic cycle. The pre-formed structure of the loop exposes the bases of these three essential nucleotides and positions them to interact with other RNA sequences within TR, with the reverse transcriptase or with the newly synthesized telomeric DNA strand. The functional role of this stem-loop appears to be conserved in even distantly related organisms such as yeast and ciliates.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle WA 98195-1700, USA.