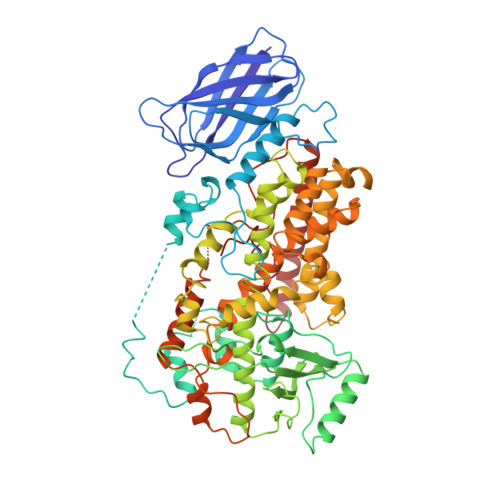
The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity.
Gillmor, S.A., Villasenor, A., Fletterick, R., Sigal, E., Browner, M.F.(1997) Nat Struct Biol 4: 1003-1009
- PubMed: 9406550
- DOI: https://doi.org/10.1038/nsb1297-1003
- Primary Citation of Related Structures:
1LOX - PubMed Abstract:
Here we report the first structure of a mammalian 15-lipoxygenase. The protein is composed of two domains; a catalytic domain and a previously unrecognized beta-barrel domain. The N-terminal beta-barrel domain has topological and sequence identify to a domain in the mammalian lipases, suggesting that these domains may have similar functions in vivo. Within the C-terminal domain, the lipoxygenase substrate binding site is a hydrophobic pocket defined by a bound inhibitor. Arachidonic acid can be docked into this deep hydrophobic pocket with the methyl end extending down into the bottom of the pocket and the acid end tethered by a conserved basic residue on the surface of the enzyme. This structure provides a unifying hypothesis for the positional specificity of mammalian lipoxygenases.
Organizational Affiliation:
Graduate Group in Biophysics, University of California, San Francisco 94143-0448, USA.