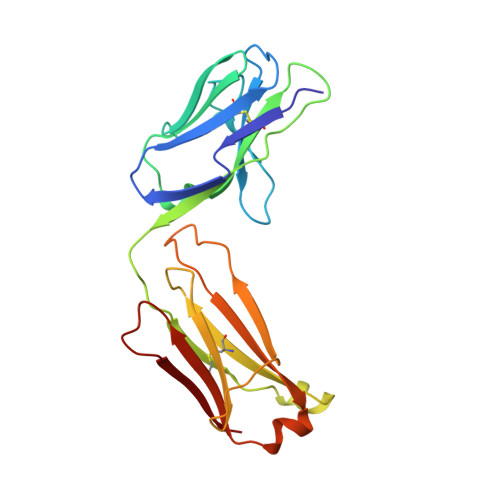
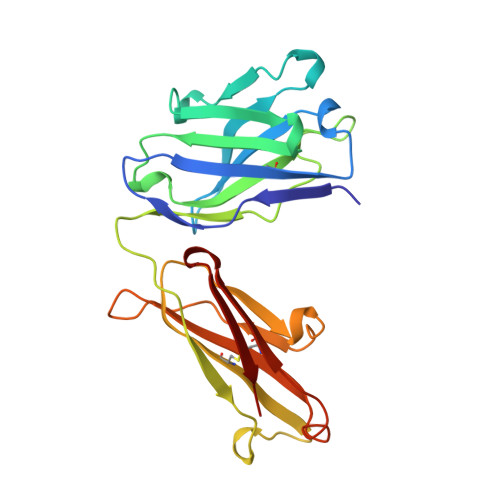
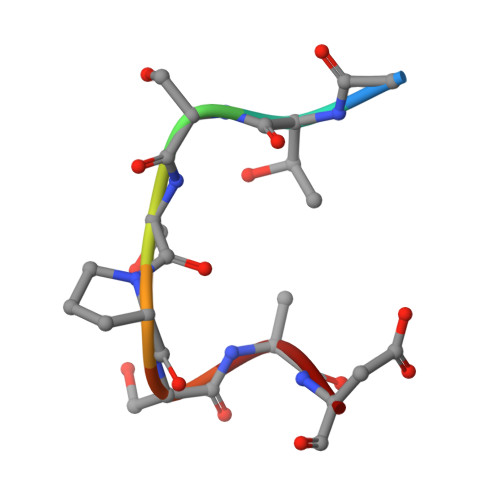
1.9 A structure of the therapeutic antibody CAMPATH-1H fab in complex with a synthetic peptide antigen.
James, L.C., Hale, G., Waldmann, H., Bloomer, A.C.(1999) J Mol Biol 289: 293-301
- PubMed: 10366506
- DOI: https://doi.org/10.1006/jmbi.1999.2750
- Primary Citation of Related Structures:
1CE1 - PubMed Abstract:
CAMPATH-1 antibodies have a long and successful history in the treatment of leukaemia, autoimmune disease and transplant rejection. The first antibody to undergo "humanisation", CAMPATH-1H, permits treatment with limited patient anti-globulin response. It recognises the CD52 antigen which is a small glycosylphosphatidylinositol(GPI)-anchored protein expressed on lymphocytes and mediates cell depletion. We present the 1.9 A structure of the CAMPATH-1H Fab complexed [corrected] with an analogue of the antigenic determinant of CD52. Analysis of the CAMPATH-1H binding site reveals that in contrast to most antibodies CDR L3 plays a dominant role in antigen binding. Furthermore CDR H3, which is essential for effective antigen recognition in most antibodies, participates in only two main-chain interactions in CAMPATH-1H. The CAMPATH-1H binding site is highly basic; ionic interaction with the enthanolamine phosphate of the CD52 GPI anchor has long been hypothesised to be important in antigen binding. The structure reveals a number of important specific ionic interactions, including Lys53H but not Lys52bH as had previously been suggested. Prolonged treatment with CAMPATH-1H can lead to patient anti-idiotype responses which may be exacerbated by the unusually high number of basic residues in the antibody. This suggests that a strategy where redundant basic residues are replaced with neutral counterparts may be effective in further reducing the immunogenicity of this versatile and widely used antibody.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 2QH, UK.